티스토리 뷰
Bioinformatics
Bioinformatics tool | GGDC, Genome-to-Genome Distance Calculator
Chloe A_Choe 2020. 1. 16. 15:17https://ggdc.dsmz.de/ggdc.php#
https://ggdc.dsmz.de/ggdc.php
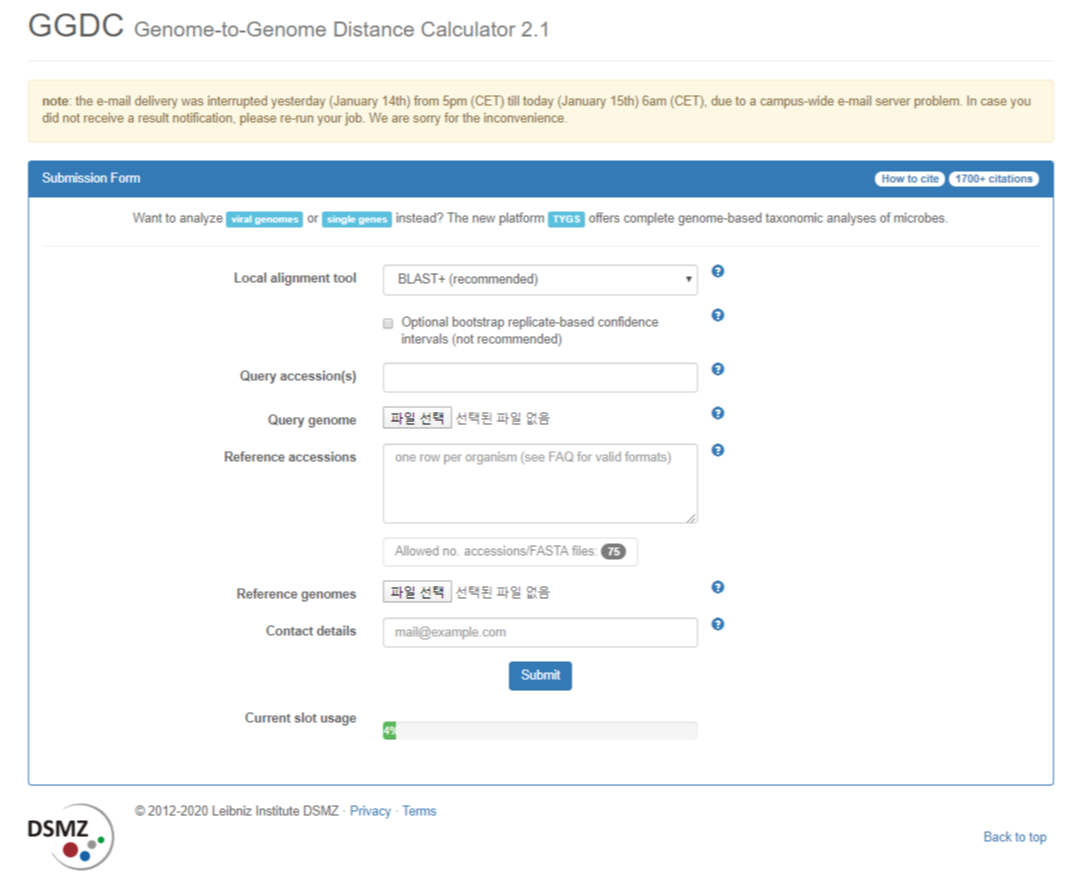
note: the e-mail delivery was interrupted yesterday (January 14th) from 5pm (CET) till today (January 15th) 6am (CET), due to a campus-wide e-mail server problem. In case you did not receive a result notification, please re-run your job. We are sorry for t
ggdc.dsmz.de
Genoem to Genome Distance Calculator(GGDC)
- Genome 간의 거리를 간편하게 계산해주는 웹 프로그램
- Query 시퀀스 파일을 직접 선택해도 되고, NCBI accession no. 를 입력해주어도 됨
- Reference 입력도 Query 입력과 동일
- 결과를 받아볼 email을 입력하면 분석이 끝나는대로 결과를 받아볼 수 있음
- 결과에서 총 3개의 formula를 제공하는데 그중에서 Formula 2를 추천함(그 이유는 아래 첨부)
In many cases the three distinct formulae give almost identical results regarding DDH. For instance, for two E. coli genomes used as test data, with BLAST+ we obtain 74.80% ± 3.80, 76.70% ± 2.87 and 77.80% ± 3.28, respectively. But for other genomes the results can be distinct, and sometimes even different regarding the 70% boundary. This is not caused by an error in the calculations but by the three distance formulae exploring distinct aspects of genome evolution.
All models for estimating DDH from intergenomic distances have been inferred separately, and all formulae yielded very high correlations. Whereas the test dataset used was larger than in any previous studies, we were unable to prefer a certain distance formula on the basis of the model-building results alone when using the test dataset. This does not mean, however, that other criteria would not yield stronger differences between the formulae. For instance, formula 2 is the only one that can be used with incompletely sequenced genomes. But there are other reasons why we recommend formula 2.
From a biological point of view, if all genomes have been sequenced completely (have "Finished" status), or almost so, but formula 1 yields much higher DDH similarities than formula 2, this indicates that the two strains changed comparatively little in gene content but comparatively strongly regarding the gene sequences. Depending on the kind of organisms and on the selection pressures under which they evolved, this might not be an unreasonable scenario, even though many strains within the same species differ considerably in their gene content. For instance, two strains might differ more or less only in the presence of a plasmid (see FAQ entry). This alone might be a reason for preferring formula 2, but the other two formulae potentially provide valuable additional biological information and hence are included in the results.
'Bioinformatics' 카테고리의 다른 글
공지사항
최근에 올라온 글
최근에 달린 댓글
- Total
- Today
- Yesterday
링크
TAG
- plot
- visualizing
- Python
- data
- 코로나바이러스
- 2진수
- coronavirus
- Heatmap
- 숫자
- covid
- format
- Cast
- 파이썬
- 데이터
- Visualization
- comma
- 팟빵
- Excel
- geom_line
- for loop
- 엑셀
- hist
- SEQ
- r
- BIOINFORMATICS
- Coding
- geom_bar
- Order
- Command
- RStudio
| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 |
글 보관함
